CAMP: Computational Anatomy and Medical imaging using PyTorch
Blake E. Zimmerman
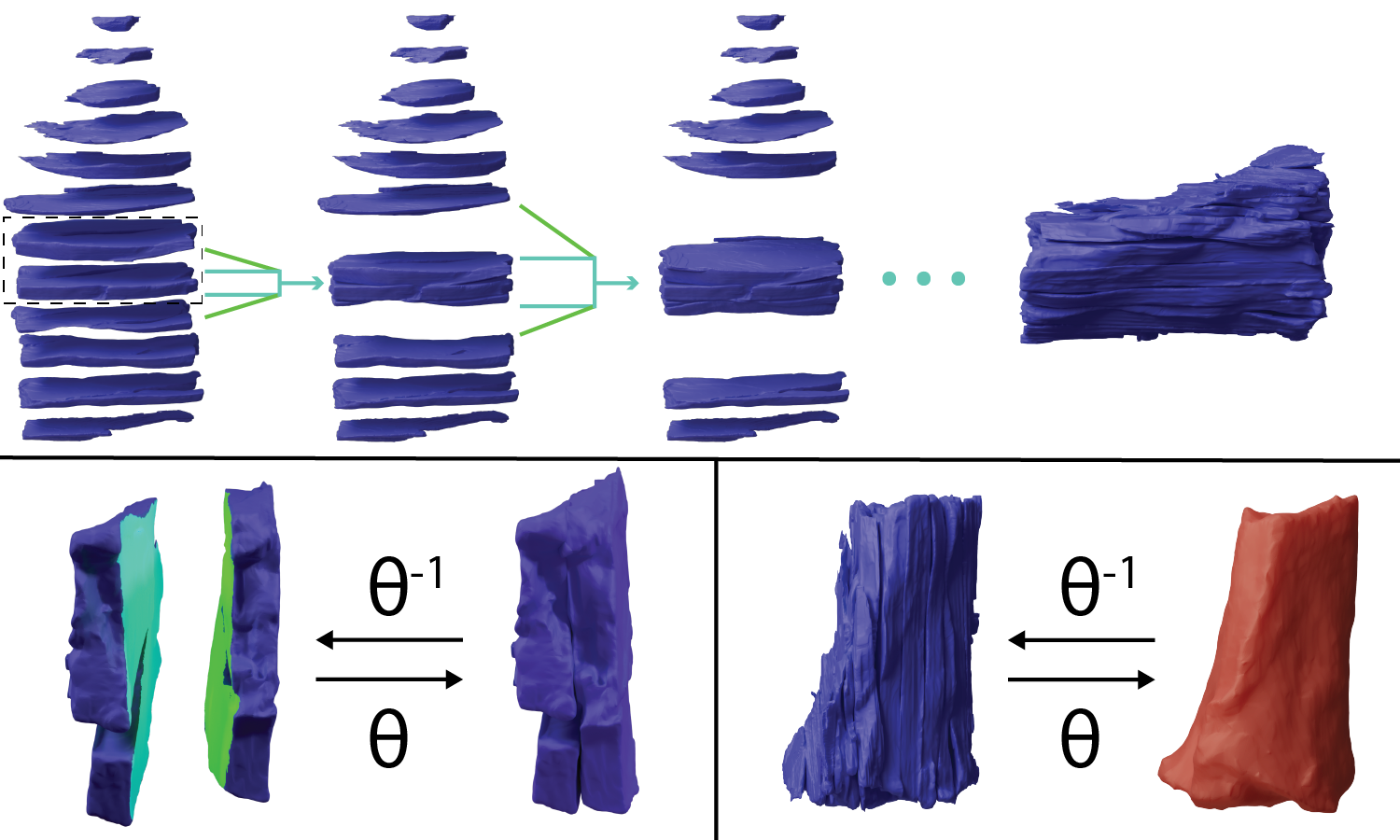
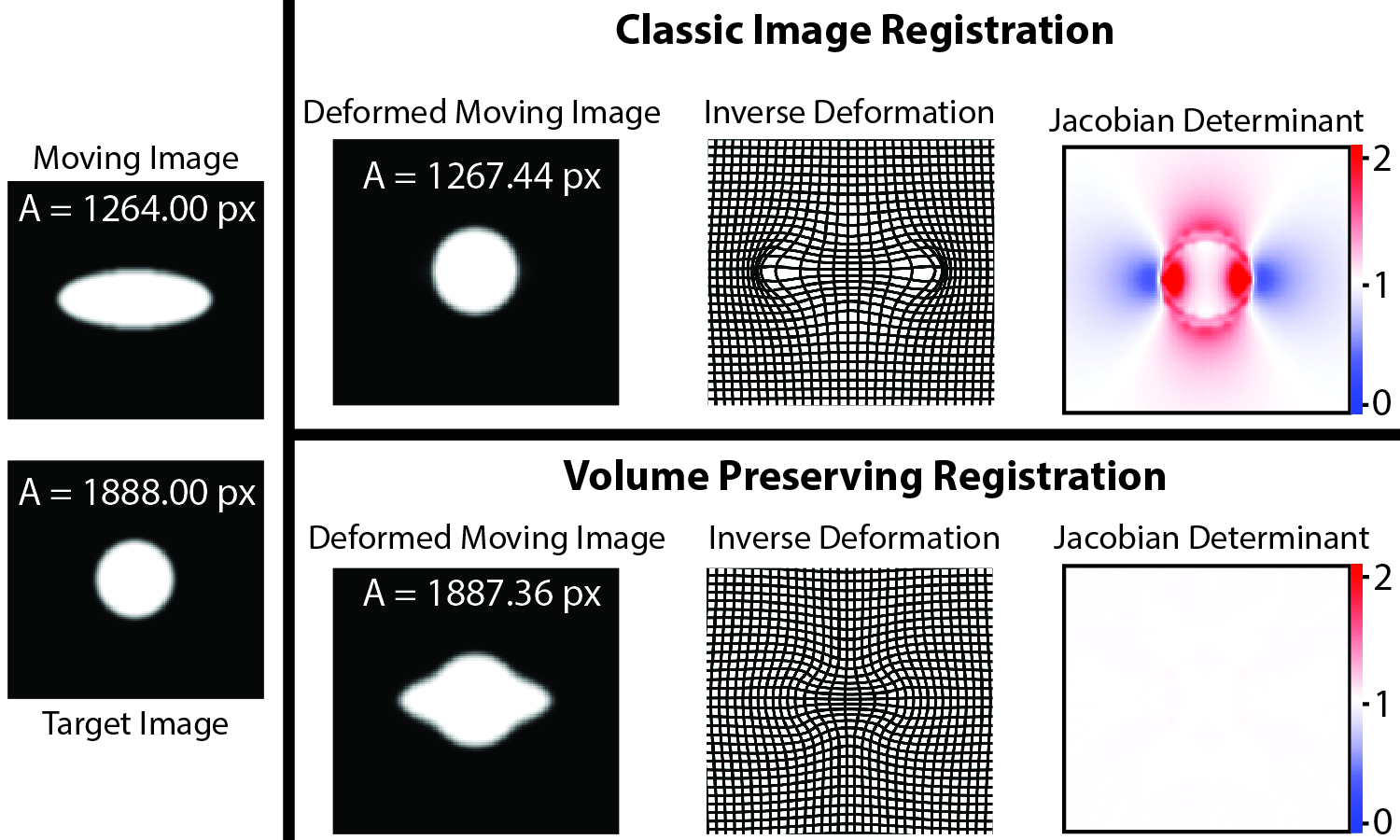
The CAMP repository was developed to leverage the memory management and computational efficiency of PyTorch while incorporating a unifying coordinate system for easy integration with medical imaging. For this repository, I wrote core techniques, including data ITK-based i/o, as building blocks other research projects. I implemented and contributed GPU-accelerated implementations of medical image registration algorithms as well as developed 3D deformable surface registration for triangular mesh objects (left image). Throughout the development of CAMP, I documented functions and examples, including coordinate system conventions for medical images. Finally, I implemented volume-preserving deformable registration to reflect physiological tissue deformations for one of our publications (right image). This code has been directly used in two of my first author publications:
- Zimmerman, Blake, Sara L. Johnson, Henrik A. Odéen, Jill E. Shea, Markus D. Foote, Nicole S. Winkler, Sarang C. Joshi, and Allison H. Payne "Learning Multiparametric Biomarkers for Assessing MR-Guided Focused Ultrasound Treatments." IEEE Transactions on Biomedical Engineering (2020).
- Zimmerman, Blake, Sara L. Johnson, Henrik A. Odéen, Jill E. Shea, Rachel E. Factor, Sarang C. Joshi, and Allison H. Payne "Histology to 3D In Vivo MR Registration for Volumetric Evaluation of MRgFUS Treatment Assessment Biomarkers." Submitted (2021).